Adleberg, Jason, Amr Wardeh, Florence X Doo, Brett Marinelli, Tessa S Cook, David S Mendelson, and Alexander Kagen. 2022. “Predicting Patient Demographics from Chest Radiographs with Deep Learning.” Journal of the American College of Radiology 19 (10): 1151–61.
Apeles, Linda. 2022. “Race-Based Prescribing for Black People with High Blood Pressure Shows No Benefit.” Patient Care.
Cerdeña, Jessica P, Marie V Plaisime, and Jennifer Tsai. 2020. “From Race-Based to Race-Conscious Medicine: How Anti-Racist Uprisings Call Us to Act.” The Lancet 396 (10257): 1125–28.
Cheng, Ching-Yu, WH Linda Kao, Nick Patterson, Arti Tandon, Christopher A Haiman, Tamara B Harris, Chao Xing, et al. 2009. “Admixture Mapping of 15,280 African Americans Identifies Obesity Susceptibility Loci on Chromosomes 5 and x.” PLoS Genetics 5 (5): e1000490.
Freedman, Matthew L, Christopher A Haiman, Nick Patterson, Gavin J McDonald, Arti Tandon, Alicja Waliszewska, Kathryn Penney, et al. 2006. “Admixture Mapping Identifies 8q24 as a Prostate Cancer Risk Locus in African-American Men.” Proceedings of the National Academy of Sciences 103 (38): 14068–73.
Garbin, Christian, Pranav Rajpurkar, Jeremy Irvin, Matthew P Lungren, and Oge Marques. 2021. “Structured Dataset Documentation: A Datasheet for CheXpert.” arXiv Preprint arXiv:2105.03020.
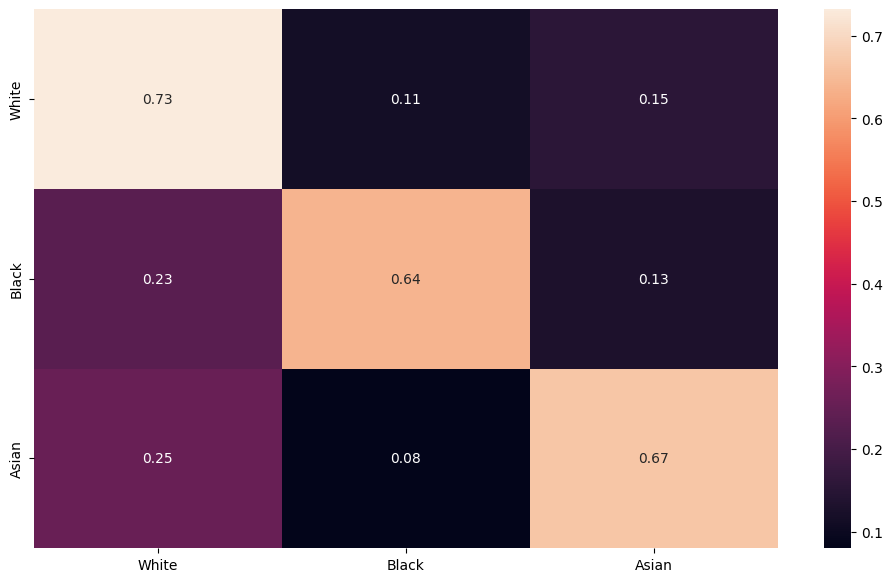
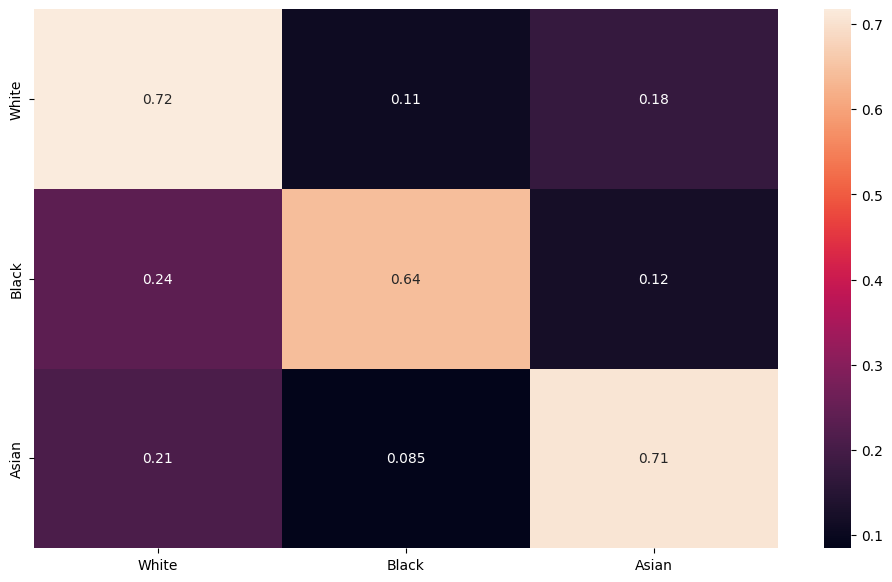
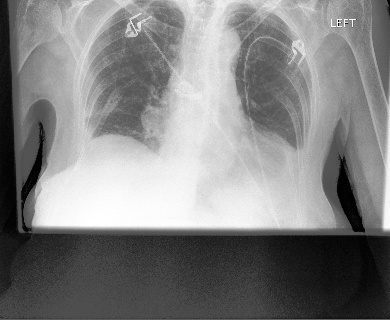
Gichoya, Judy Wawira, Imon Banerjee, Ananth Reddy Bhimireddy, John L Burns, Leo Anthony Celi, Li-Ching Chen, Ramon Correa, et al. 2022. “AI Recognition of Patient Race in Medical Imaging: A Modelling Study.” The Lancet Digital Health 4 (6): e406–14.
Irvin, Jeremy, Pranav Rajpurkar, Michael Ko, Yifan Yu, Silviana Ciurea-Ilcus, Chris Chute, Henrik Marklund, et al. 2019. “Chexpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison.” In Proceedings of the AAAI Conference on Artificial Intelligence, 33:590–97. 01.
Maglo, Koffi N, Tesfaye B Mersha, and Lisa J Martin. 2016. “Population Genomics and the Statistical Values of Race: An Interdisciplinary Perspective on the Biological Classification of Human Populations and Implications for Clinical Genetic Epidemiological Research.” Frontiers in Genetics 7: 22.
Reich, David, Nick Patterson, Philip L De Jager, Gavin J McDonald, Alicja Waliszewska, Arti Tandon, Robin R Lincoln, et al. 2005. “A Whole-Genome Admixture Scan Finds a Candidate Locus for Multiple Sclerosis Susceptibility.” Nature Genetics 37 (10): 1113–18.
Seyyed-Kalantari, Laleh, Haoran Zhang, Matthew BA McDermott, Irene Y Chen, and Marzyeh Ghassemi. 2021. “Underdiagnosis Bias of Artificial Intelligence Algorithms Applied to Chest Radiographs in Under-Served Patient Populations.” Nature Medicine 27 (12): 2176–82.
Tang, Hua, Tom Quertermous, Beatriz Rodriguez, Sharon LR Kardia, Xiaofeng Zhu, Andrew Brown, James S Pankow, et al. 2005. “Genetic Structure, Self-Identified Race/Ethnicity, and Confounding in Case-Control Association Studies.” The American Journal of Human Genetics 76 (2): 268–75.
Vergara, Candelaria, Luis Caraballo, Dilia Mercado, Silvia Jimenez, Winston Rojas, Nicholas Rafaels, Tracey Hand, et al. 2009. “African Ancestry Is Associated with Risk of Asthma and High Total Serum IgE in a Population from the Caribbean Coast of Colombia.” Human Genetics 125: 565–79.
Vyas, Darshali A, Leo G Eisenstein, and David S Jones. 2020. “Hidden in Plain Sight—Reconsidering the Use of Race Correction in Clinical Algorithms.” New England Journal of Medicine. Mass Medical Soc.
Yi, Paul H, Jinchi Wei, Tae Kyung Kim, Jiwon Shin, Haris I Sair, Ferdinand K Hui, Gregory D Hager, and Cheng Ting Lin. 2021. “Radiology ‘Forensics’: Determination of Age and Sex from Chest Radiographs Using Deep Learning.” Emergency Radiology 28: 949–54.